Innovation comes only from the assault on the unknown.”
Research
In vivo functional genetics of noncoding genomic regions
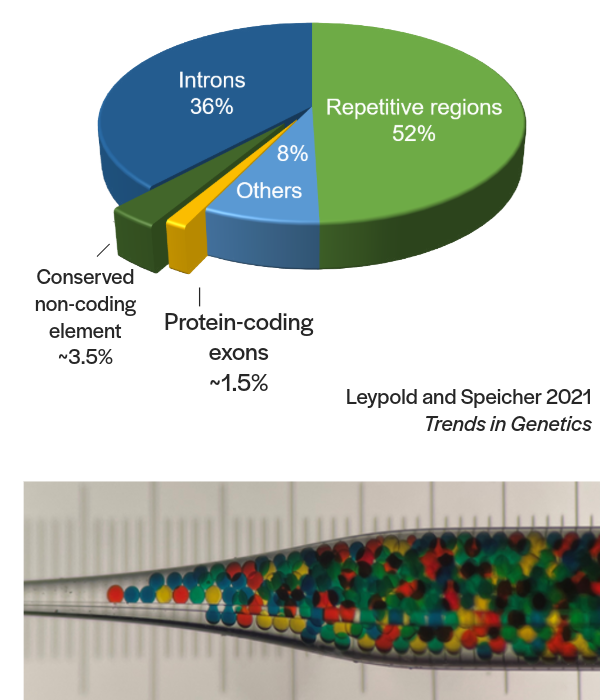 Noncoding genomic regions constitute a vast majority of the human genome. Despite their presumed importance, direct evidence linking a noncoding genomic region to its function, especially in an in vivo model system, has been limited.
Noncoding genomic regions constitute a vast majority of the human genome. Despite their presumed importance, direct evidence linking a noncoding genomic region to its function, especially in an in vivo model system, has been limited.
We are using comparative genomics approaches to identify conserved noncoding regions. Our rationale is that regions that are conserved through millions of years of evolution are likely functional. We are interrogating the functions of these regions by targeted perturbation in zebrafish using Multiplexed Intermixed CRISPR Droplets (MIC-Drop), a unique platform that enables large-scale CRISPR genetic screens to be conducted in a whole animal model system.
We are using a combination of morphological, behavioral and molecular phenotyping approaches to get a comprehensive understanding of the functions of these noncoding regions. Our lab intends to do deeper mechanistic characterization of how these noncoding regions regulate the observed developmental phenotypes.
Novel technologies for targeted and efficient genetic perturbation in zebrafish and deeper phenotypic characterization
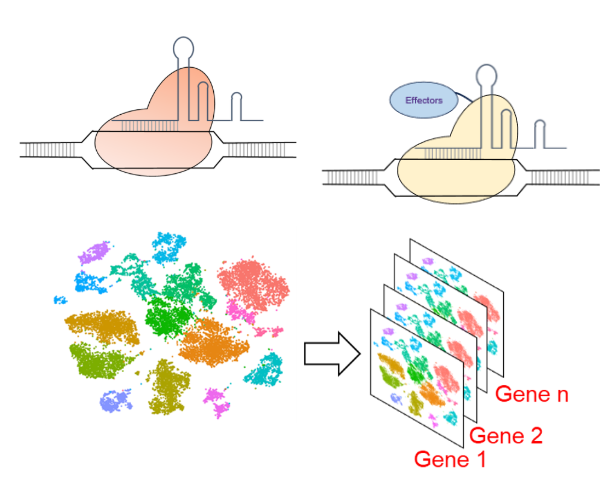
Functional characterization of the noncoding genome has lagged because of:
- A dearth of tools for efficient and targeted perturbation of noncoding regions in vivo.
- The lack of technologies for deeper characterization of mutant phenotypes.
We are developing technologies on both fronts. Even though CRISPR-Cas9 has made targeted gene knockout in zebrafish rapid and facile, perturbing noncoding genomic region presents new sets of challenges. We are developing CRISPR-based technologies that enable reliable and efficient targeting of noncoding genomic regions in zebrafish.
In addition, we are developing novel technologies that will enable a more comprehensive understanding of mutant phenotypes. This includes high-throughput morphometric assessments using state-of-the-art high-content imaging platforms, behavioral fingerprinting, and molecular characterization using single-cell RNA sequencing.
Progress in science depends on new techniques, new discoveries, and new ideas, probably in that order. ”
Disease modeling in zebrafish and mechanistic characterization
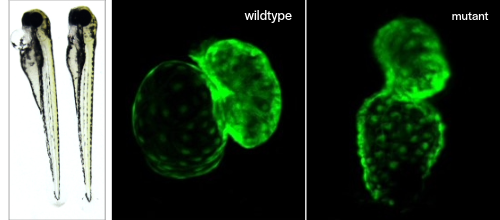
A tractable genome and conserved developmental pathways make zebrafish a great model system for modeling human diseases and revealing the mechanisms of action. From prior genetic screens, we have identified several candidate genes, the loss-of-function of which results in cardiovascular defects in zebrafish.
Our lab is using standard developmental biology approaches to understand the mechanisms underlying the observed phenotype.
Despite the diversity of organisms, we share a lot in common [...] studies of simple non-human organisms can provide crucial breakthroughs important for the understanding, prevention and cure of human disease.”