siRNAs Based on Huntington Trinucleotide Repeats Are Highly Toxic to All Cancer Cells
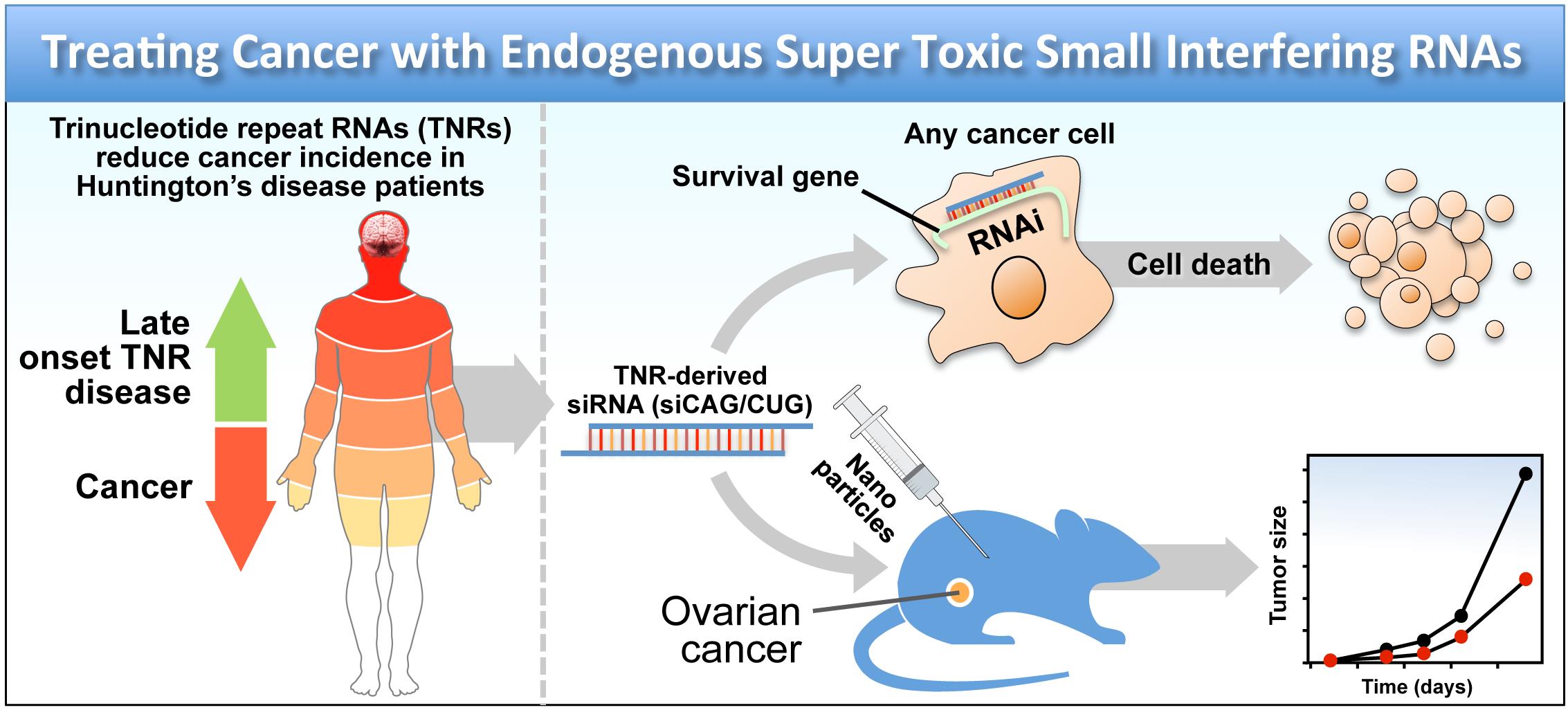
The Peter Lab previously identified toxic small interfering RNAs that killed all cancer cells in a way that they could not become resistant to (1, 2). They have now identified a new class of related molecules that are about 100 times more toxic to cancer cells than the previously described ones (3). These super toxic siRNAs are based on naturally occurring RNAs in patients with certain neurological diseases including Huntington's disease (HD). HD patients suffer from a terminal disease caused by amplification of certain repeat sequences in the human genome. The repeat module that is amplified in these patients is the trinucleotide sequence C-A-G. While the patients suffer from this disease, they have lower cancer rates than the general population. The Peter Lab has now found that these CAG sequences at the RNA level are super toxic to all cancer cells when expressed as siRNAs (siCAG/CUG) and can in fact be used to treat cancer, providing an explanation for why Huntington's disease patients have less cancer. Treatment of advanced metastasizing cancer is a big unsolved problem. The new treatment based on CAG repeats so far has not shown any toxicity in mice and no development of resistance to the treatment has been observed. The plan is to further develop this treatment for clinical use in cancer patients. While HD patients suffer from a debilitating disease, it usually does not manifest itself before the age of 40. A short term treatment of cancer with siCAG/CUG for a few weeks may therefore be possible without causing the neurological issues that HD patients suffer from. Interestingly, while CAG trinucleotide repeat (TNR) expansion patients show reduced cancer incidence CUG based diseases such as myotonic dystrophy type I (DM1) show increased cancer incidences. The Peter Lab recently proposed the TNRs in the genome as being part of an anti cancer mechanism and provided a new hypothesis to explain the opposite effects of CAG and CUG TNRs on cancer risk (4).
References
- Putzbach, W., Gao, Q.Q., Patel, M., van Dongen, S., Haluck-Kangas, A., Sarshad, A.A., Bartom, E., Kim, K.Y., Scholtens, D.M., Hafner, M., Zhao, J.C., Murmann, A.E., and Peter, M.E. (2017) Many si/shRNAs can kill cancer cells by targeting multiple survival genes through an off-target mechanism. eLife, 6, e29702.
- Putzbach, W., Gao, Q.Q., Patel, M., Haluck-Kangas, A., Murmann, A.E. and Peter, M.E. (2018) DISE - An RNAi Off-Target Effect that Kills Cancer Cells. Trends in Cancer, 4, 10-19.
- Murmann, A.E., Putzbach, W., Gao, Q., Patel, M., Bartom, T. E., Law, C., Bridgeman, B., Chen, S., McMahon, K.M., Thaxton, C.S., and Peter, M.E. (2018) Small interfering RNAs based on huntington trinucleotide repeats are highly toxic to cancer cells. EMBO Rep, 19, e45336.
- Murmann, A.E., Yu, J., Opal, P. and Peter, M.E. (2018) Trinucleotide repeat expansion diseases, RNAi and cancer, Trends in Cancer, 4, 684-700.