6mer Seed Toxicity - A Kill Code Embedded in the Genome
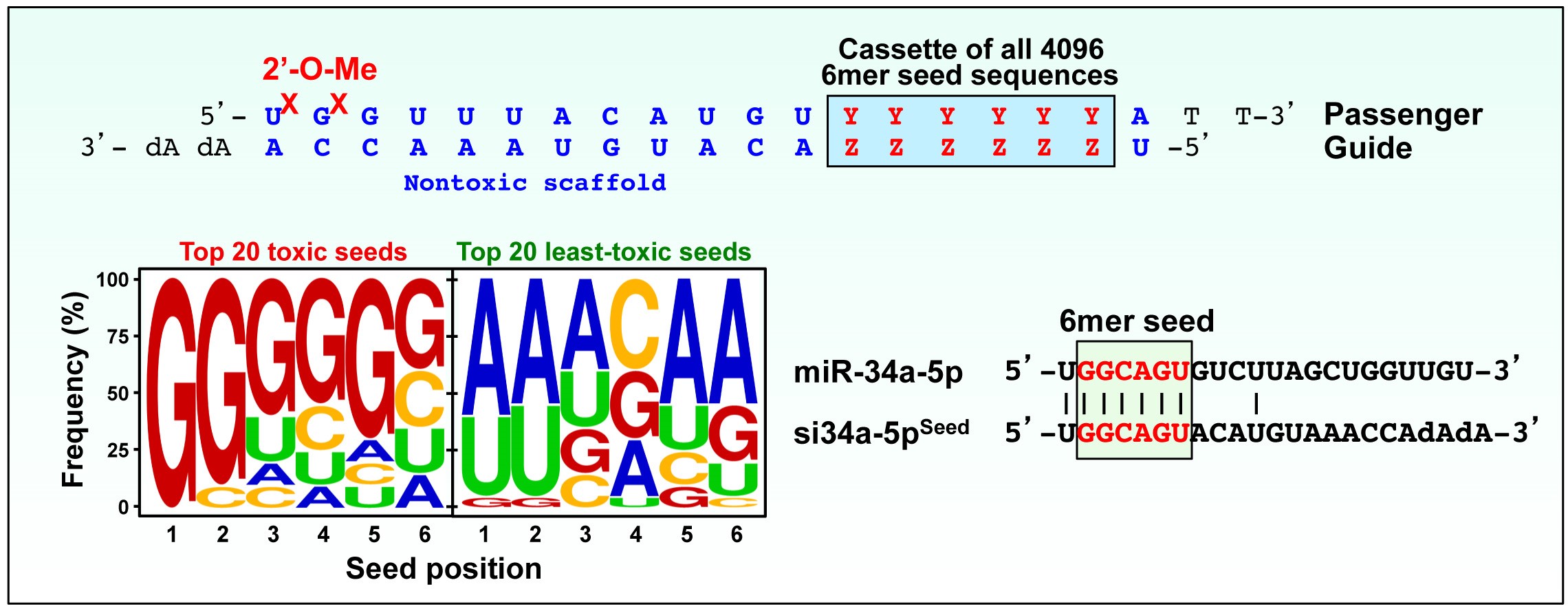
New data demonstrated that DISE induction is a promising new approach for treating cancer. Subsequent to this work the Peter Lab determined that only a 6mer seed (positions 2-7 of the guide strand of the siRNA) was necessary to induce DISE (1). This allowed to test all possible 4096 6mer seed sequences. This screen revealed that the rules of toxicity were very similar for all four tested human and mouse cell lines suggesting that 6mer seed toxicity kills cancer cells regardless of their tissue or species of origin (see 6merdb.org). The most toxic of all 6mer seeds carried two Gs at the 5' end and a C at the 3' end of the seed (2). These toxic seed containing siRNAs target in a miRNA-like fashion a large set of critical survival genes through C-rich seed matches located in their 3'UTRs validating the DISE concept.
Many tumor suppressive miRNAs such as miR-34a-5p but none of the established oncogenic miRNAs contain G-rich 6mer seeds and most of toxicity of the master tumor suppressive miRNA miR-34a-5p comes from its 6mer seed sequence (2). For most miRNAs the more abundant mature form corresponds to the arm that contains the less toxic seed. In contrast, for major tumor suppressive miRNAs, the mature miRNA is derived from the arm that harbors the more toxic seed. The data allow to conclude that while most miRNAs over >800 Million years have evolved to avoid targeting survival and housekeeping genes, certain tumor suppressive miRNAs function to kill cancer cells through a toxic G-rich 6mer seed targeting the 3'UTR of survival genes.
Another finding that came out of this study is that many chemo therapeutic/genotoxic drugs kill cancer cells at least in part by triggering DISE (2), through inducing the production of a wave of small RNAs that are toxic to cells through 6mer seed toxicity.
Most recently the Peter Lab demonstrated that not only CD95L derived siRNAs but the entire mRNA of CD95L is toxic to cancer cells and kills through induction of DISE (3). 22 distinct regions within the CD95L CDS were identified that can give rise to small RISC-bound RNAs. Finally, about 3% of the coding genes in the human genome were found to be processed in this way under conditions of low miRNA expression typical for advanced cancers. These genes include almost exclusively genes that are required for protein translation and cell growth (3). The data suggest that DISE is a process that is involved in fighting cancer and that large parts of the coding genome can be processed and affect cell fate through RNAi.
References
- Putzbach, W, Gao, QQ, Patel, M, van Dongen, S, Haluck-Kangas, A, Sarshad, AA, Bartom, E, Kim, KY, Scholtens, DM, Hafner, M, Zhao, JC, Murmann, AE, and Peter, ME (2017) Many si/shRNAs can kill cancer cells by targeting multiple survival genes through an off-target mechanism. eLife, 6, e29702.
- Gao, Q.Q., Putzbach, W.E., Murmann, A.E., Chen, S., Sarshad, A.A., Peter, J.M., Bartom, E.T., Hafner, M. and Peter, M.E. (2018) 6mer seed toxicity in tumor suppressive microRNAs. Nature Comm, 9, 4504.
- Putzbach, W., Haluck-Kangas, A., Gao, Q.Q., Sarshad, A.A., Bartom, E. T., Stults, A.M., Qadir, A.S., Hafner, M. and Peter, M.E. (2018) CD95/Fas ligand mRNA is toxic to cells. eLife, 7, e38621.