Research
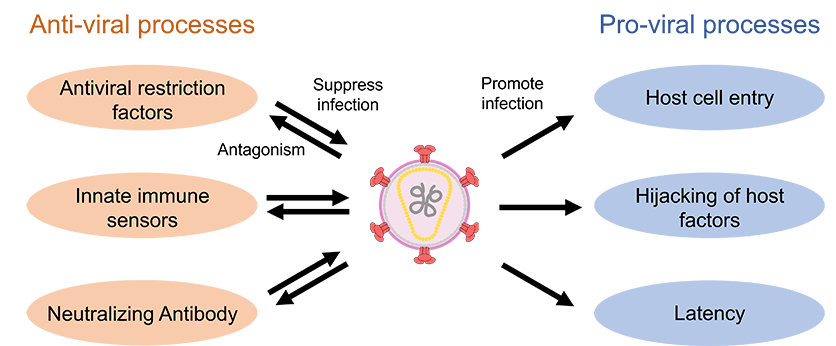
When viruses infect host cells, they establish a complex network of interactions with host factors to transform the cells into a fully habitable environment. Such interaction includes the initial attachment of the virus to the target cell surface through specific cell-entry receptors, hijacking of host cellular machinery for productive viral replication, and the evasion of host immunity through viral protein antagonists. For example, just a handful of genes encoded by human immunodeficiency virus (HIV) multitask to facilitate the infection, which involves numerous interactions with the host proteins and nucleic acids. Targeting virus-host interactions within infected cells represents an important avenue for the future development of versatile and adaptable therapeutics for many types of viral infections. To this end, comprehensive structural and functional information on the essential interactions between viral components and host factors is essential.
Our long-term goal is to yield atomic-level molecular insights into viral infection, host immune defense, and virus-host interaction by using cryo-electron microscopy (cryo-EM) combined with biochemical and virological functional approaches. Additionally, we aim to identify unknown host factors associated with the viral proteins of interest by using cryo-EM-based structural proteomics.
Current Projects
Project 1: Molecular basis of HIV accessary protein complexes 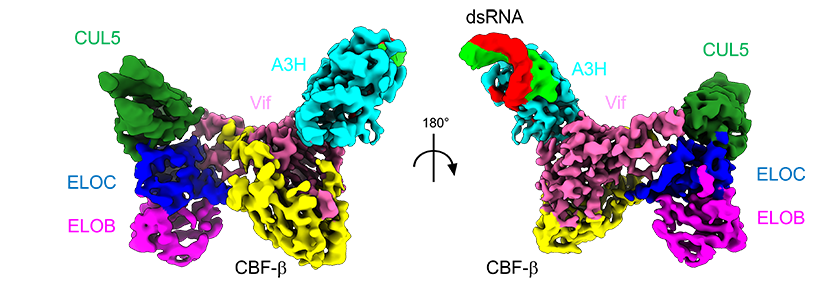
HIV-1 possesses a compact genome of 9.7 kb, encoding just nine genes. In addition to the typical retrovirus structural and enzymatic genes gag, pol, and env, the HIV-1 genome contains four accessory genes vpr, vif, vpu, and nef. The HIV accessory proteins are not directly involved in the viral genome replication, virus assembly, or maturation, but they play important roles in maintaining the infectivity of the virus, particularly in the late stages of the HIV lifecycle. Typically, they promote the virus infectivity by intercepting essential host processes such as antiviral immune response. For example, Vif hijacks the host Cul5 E3 ubiquitin ligase complex to degrade human APOBEC3 antiviral factors in T-cells. Because of this, the deletion of vif gene from the HIV genome results in severely compromised infectivity of the virus in T-lymphoblast CEM cell lines. While the role of Vpr in HIV replication remains enigmatic, studies indicated that Vpr is required for viral replication in macrophages. This underscores the potential of targeting these accessory proteins in mitigating HIV pathogenicity.
Building upon our previous Vif-APOBEC3 complex studies, we seek to comprehensively delineate the molecular mechanisms governing the interactions of the HIV accessory proteins and their host targets. Defining such virus-host molecular interactions would provide crucial insights into the intricate strategy that viruses adopt to combat host anti-viral factors and to exploit pro-viral factors during viral infection.
Project 2: Extended assembly and interactions of coronavirus replication-transcription complex
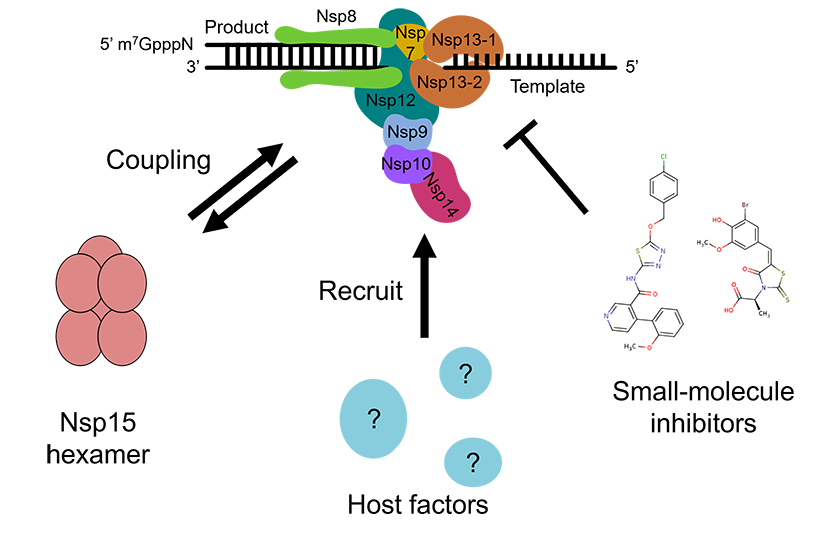
The replication–transcription complex (RTC) is the central machinery that performs the replication of single-stranded RNA genome in coronaviruses, such as SARS-CoV-2. The RTC is composed of multiple non-structural proteins (Nsps), including the RNA-dependent RNA polymerase (Nsp12), helicase (Nsp13), and several associated cofactors (e.g., Nsp7, Nsp8, Nsp9, Nsp10, Nsp14, and Nsp16), which assemble into a large, multifunctional molecular machine responsible for RNA synthesis, proofreading, and 5′-end capping.
Coronavirus Nsp15 endoribonuclease processes the viral RNA for innate immune evasion, and functionally couples with the core RTC. However, the mechanism and timing of this coordination remain elusive. Moreover, emerging data indicates that the RTC incorporates multiple host proteins that assist in viral replication during infection, yet the composition and architecture of these extended assemblies are not fully defined.
In this project, we aim to reconstitute or isolate the extended SARS-CoV-2 RTC with or without its host factors in near-physiological context and characterize the complex through cryo-EM and functional approaches. By visualizing the molecular organization and regulatory mechanisms of this complex, we aim to reveal new insights into coronavirus replication and identify potential inhibitor sites that could disrupt RTC assembly or function for the future development of broad-spectrum antivirals.